Signed-off-by: Rafael Ravedutti <rafaelravedutti@gmail.com>
MD-Bench
MD-Bench is a toolbox for the performance engineering of short-range force calculation kernels on molecular-dynamics applications. It aims at covering all available state-of-the-art algorithms from different community codes such as LAMMPS and GROMACS.
Apart from that, many tools to study and evaluate the in-depth performance of such kernels on distinct hardware are offered, like gather-bench, a standalone benchmark that mimics the data movement from MD kernels and the stubbed force calculation cases that focus on isolating the impacts caused by memory latencies and control flow divergence contributions in the overall performance.
| Verlet Lists | GROMACS MxN | Stubbed cases |
|---|---|---|
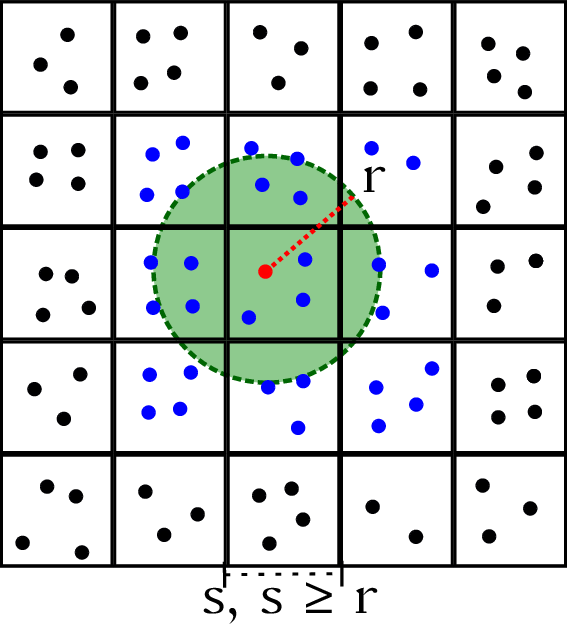 |
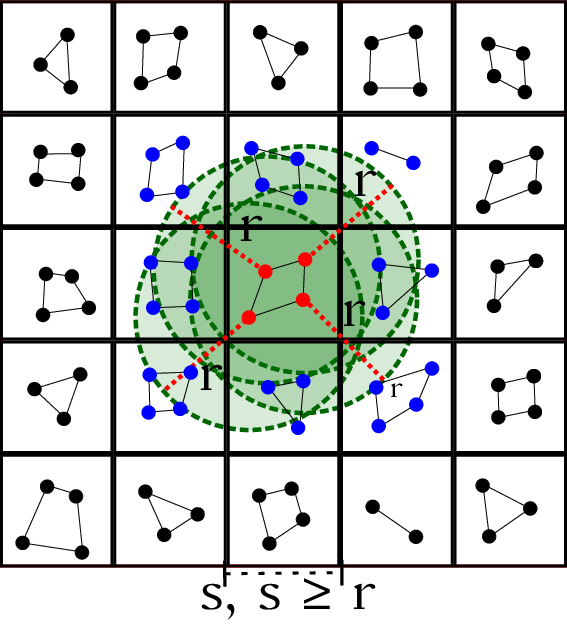 |
 |
Build instructions
Properly configure your building by changing config.mk file. The following options are available:
- TAG: Compiler tag (available options: GCC, CLANG, ICC, ONEAPI, NVCC).
- ISA: Instruction set (available options: SSE, AVX, AVX_FMA, AVX2, AVX512).
- MASK_REGISTERS: Use AVX512 mask registers (always true when ISA is set to AVX512).
- OPT_SCHEME: Optimization algorithm (available options: lammps, gromacs).
- ENABLE_LIKWID: Enable likwid to make use of HPM counters.
- DATA_TYPE: Floating-point precision (available options: SP, DP).
- DATA_LAYOUT: Data layout for atom vector properties (available options: AOS, SOA).
- ASM_SYNTAX: Assembly syntax to use when generating assembly files (available options: ATT, INTEL).
- DEBUG: Toggle debug mode.
- EXPLICIT_TYPES: Explicitly store and load atom types.
- MEM_TRACER: Trace memory addresses for cache simulator.
- INDEX_TRACER: Trace indexes and distances for gather-md.
- COMPUTE_STATS: Compute statistics.
Configurations for LAMMPS Verlet Lists optimization scheme:
- ENABLE_OMP_SIMD: Use omp simd pragma on half neighbor-lists kernels.
- USE_SIMD_KERNEL: Compile kernel with explicit SIMD intrinsics.
Configurations for GROMACS MxN optimization scheme:
- USE_REFERENCE_VERSION: Use reference version (only for correction purposes).
- XTC_OUTPUT: Enable XTC output.
- HALF_NEIGHBOR_LISTS_CHECK_CJ: Check if j-clusters are local when decreasing the reaction force.
Configurations for CUDA:
- USE_CUDA_HOST_MEMORY: Use CUDA host memory to optimize host-device transfers.
When done, just use make to compile the code.
You can clean intermediate build results with make clean, and all build results with make distclean.
You have to call make clean before make if you changed the build settings.
Usage
Use the following command to run a simulation:
./MD-Bench-<TAG>-<OPT_SCHEME> [OPTION]...
Where TAG and OPT_SCHEME correspond to the building options with the same name.
Without any options, a Copper FCC lattice system with size 32x32x32 (131072 atoms) over 200 time-steps using the Lennard-Jones potential (sigma=1.0, epsilon=1.0) is simulated.
The default behavior and other options can be changed using the following parameters:
-p <string>: file to read parameters from (can be specified more than once)
-f <string>: force field (lj or eam), default lj
-i <string>: input file with atom positions (dump)
-e <string>: input file for EAM
-n / --nsteps <int>: set number of timesteps for simulation
-nx/-ny/-nz <int>: set linear dimension of systembox in x/y/z direction
-r / --radius <real>: set cutoff radius
-s / --skin <real>: set skin (verlet buffer)
--freq <real>: processor frequency (GHz)
--vtk <string>: VTK file for visualization
--xtc <string>: XTC file for visualization
Examples
TBD
Citations
Rafael Ravedutti Lucio Machado, Jan Eitzinger, Harald Köstler, and Gerhard Wellein: MD-Bench: A generic proxy-app toolbox for state-of-the-art molecular dynamics algorithms. Accepted for PPAM 2022, the 14th International Conference on Parallel Processing and Applied Mathematics, Gdansk, Poland, September 11-14, 2022. PPAM 2022 Best Paper Award. Preprint: arXiv:2207.13094
Credits
MD-Bench is developed by the Erlangen National High Performance Computing Center (NHR@FAU) at the University of Erlangen-Nürnberg.